From 48 Hours to 6 Minutes: My Journey Optimizing a Pandas Reconciliation Process for Large-Scale Data
Introduction
I recently went through a wild but rewarding ride optimizing a heavy pandas-based reconciliation workflow. What started as a slow, clunky piece of code eating up over 48 hours, ended up getting polished into a lean, mean 6-minutes machine.
I wanted to share my story – not as a tutorial, but as a real-world case study from the trenches. If you’ve ever felt your pandas code was taking forever, this post is for you.
The Problem: Reconciling Huge DataFrames
I was working with two large pandas DataFrames – let’s call them df1 and df2, and to reconcile under two categories:
- Exact Match
- Tolerance-Based Match
The reconciliation keys were mostly PAN, TAN, TAXABLE_VALUE, TDS and DATE, but for the second category, TAXABLE_VALUE & TDS match allowed a tolerance window – not an exact match. The date tolerance was applicable in both categories (i.e., not a strict equality match on date)
Here’s how it looked:
- category_1: Grouped by [PAN, TAN, TAXABLE_VALUE, TDS]
- category_2: Grouped by just [PAN, TAN], since we wanted more flexibility with TAXABLE_VALUE & TDS tolerance
A Look into Data Complexity
The data wasn’t just large – it was messy and unbalanced, which made the problem more challenging:
- 3.9 million rows in one DataFrame (df1) and 770,000 rows in the other DataFrame (df2)
- Duplicate entries on both sides for the same PAN-TAN combinations
- Multiple TANs under a single PAN, not always aligned between df1 and df2
- Skewed distribution – some PAN-TAN groups had thousands of rows, others had only a few
This resulted in combinatorial explosions when matching duplicate groups – especially during tolerance-based reconciliation – and made naive multiprocessing very inefficient.
Below are samples of how the actual data looks-
df1-
| PAN | TAN | TAXABLE_VALUE | TDS | date |
| ABCDE1234F | TAN001 | 1000 | 100 | 2023-04-01 |
| ABCDE1234F | TAN001 | 1000 | 100 | 2023-04-01 |
| ABCDE1234F | TAN002 | 2000 | 200 | 2023-04-05 |
| ABCDE1234F | TAN002 | 3000 | 300 | 2023-04-10 |
| … | … | … | … | … |
| … | … | … | … | … |
df2-
| PAN | TAN | TAXABLE_VALUE | TDS | date |
| ABCDE1234F | TAN001 | 1000 | 100 | 2023-04-02 |
| ABCDE1234F | TAN001 | 999 | 101 | 2023-04-03 |
| ABCDE1234F | TAN002 | 1995 | 198 | 2023-04-06 |
| ABCDE1234F | TAN004 | 4000 | 400 | 2023-04-11 |
| … | … | … | … | … |
First Attempt
The initial implementation had two separate methods, one for each category. Each ran independently in its own multiprocessing pool. I was relying heavily on .apply(lambda…) for logic like date tolerance, and the performance was… let’s just say, not ideal.
def complex_check(row, category):
.... .... ....
time_delta = (dt.strptime(row[DATE_X], "%Y-%m-%d") - dt.strptime(row[DATE_Y], "%Y-%m-%d")).days
if not time_delta or LOWER_BOUND <= time_delta <= UPPER_BOUND:
return category
return "X"
# Here, data and df3 are derived from df1 and df2, respectively. .... .... .... data.set_index(data[KEY_INDEX], inplace=True) df3.set_index(df3[KEY_INDEX], inplace=True) df3 = pd.merge(data, df3, left_index=True, right_index=True) if df3.empty: continue .... ... .... df3[CATEGORY] = df3.apply(lambda record: complex_check(record, category), axis=1) .... .... ....
Task parallelism is driven by the unique values in the KEY_INDEX column of df1. Let’s explore this further below.
.... .... .... df1.set_index(df1[KEY_INDEX], inplace=True) df2.set_index(df2[KEY_INDEX], inplace=True) jobs = [] with Pool(processes=ie.n_processes) as pool: for key_index in df1[KEY_INDEX].unique(): data = df1[df1[KEY_INDEX] == key_index] df3 = df2[df2[KEY_INDEX] == key_index] if not data.empty and not df3.empty: jobs.append(pool.apply_async(partial(func, data, df3, category, ... .... ....))) if jobs: df = pd.concat([job.get() for job in jobs]) .... .... .... .... .... ....
Execution Time: Category 1: 46.53 hours Category 2: 1.40 hours
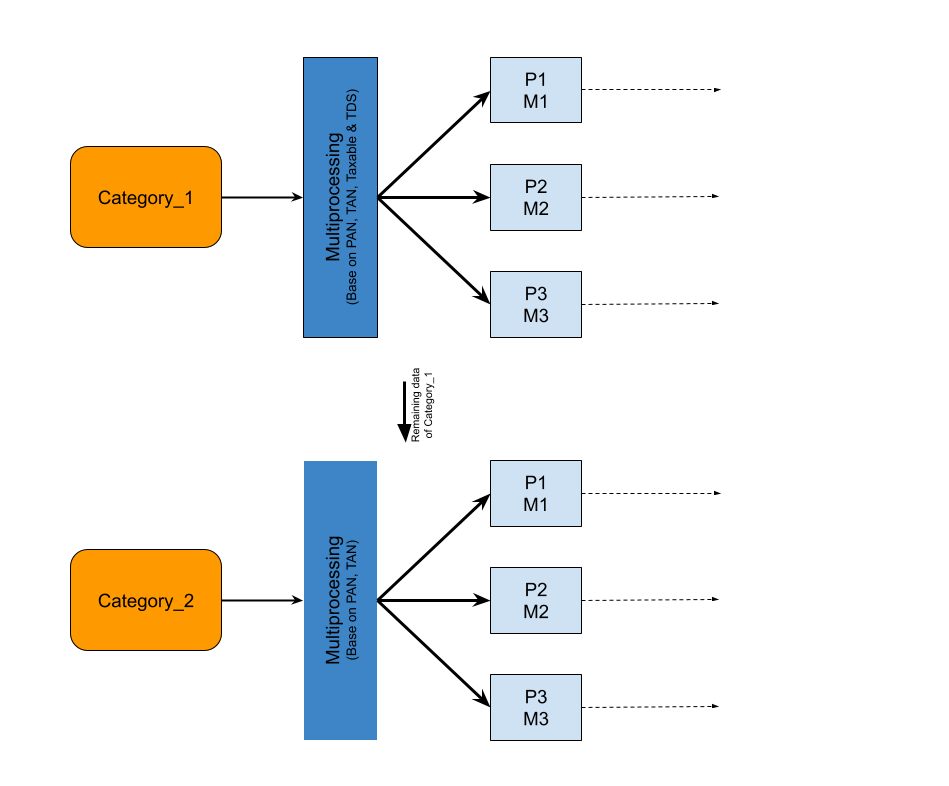
Execution Flow Diagram: Before Optimization
Refactoring & Optimizations
I knew I had to fix this. So I took a phased approach, breaking down the bottlenecks and tackling them one by one.
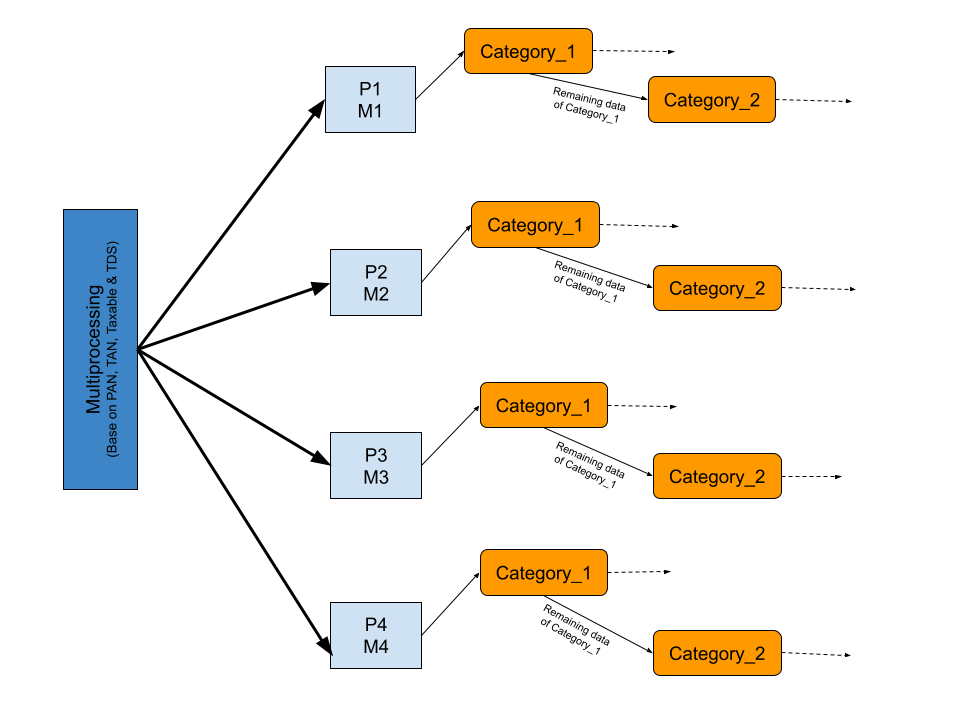
Execution Flow Diagram: After Optimization
Case 1: Merging Parallel Workflows
Problem: I was running both categories separately, duplicating effort and wasting resources.
Fix: Instead of parallelizing the categories, I parallelized each PAN-TAN combination, and then handled both categories together in each process.
# Here, we are determining how many tasks will be processed in parallel for the chunks of PAN-TAN.
.... .... ....
groups_df1 = dict(list(df1.groupby([PAN, TAN])))
groups_df2 = dict(list(df2.groupby([PAN, TAN])))
common_keys = groups_df1.keys() & groups_df2.keys()
max_workers = int(os.cpu_count() * 0.85)
manager = multiprocessing.Manager()
shared_list = manager.list()
grouped_task = [
(shared_list, key, groups_df1.pop(key), groups_df2.pop(key), ..., ..., ....) for key in common_keys
]
with ProcessPoolExecutor(max_workers=max_workers) as executor:
results = list(executor.map(process_pan_tan_chunk, grouped_task))
if results:
.... .... ....
.... .... ....
del results, grouped_task, common_keys, groups_df1, groups_df2
.... .... ....
Result: Execution time dropped from 46+ hours to 15 hours. Not amazing, but hey — progress!
Case 2: Ditching .apply() for Vectorization
Problem: I was doing too much in .apply(), especially around date tolerance.
Fix: I rewrote those pieces using pandas vectorized operations – things like broadcasting ranges and using .merge_asof() instead of row-wise checks.
# Here, Vectorized version of .apply(lambda .... ....) .... .... .... df3 = pd.merge(data, df3, left_index=True, right_index=True) if df3.empty: continue df3["delta_days"] = (pd.to_datetime(df3[DATE_X]) - pd.to_datetime(df3[DATE_Y])).dt.days df3[CATEGORY] = np.where((df3["delta_days"] >= LOWER_BOUND) & (df3["delta_days"] <= UPPER_BOUND), category, "X") .... .... ....
Result: Boom - down to 2 hours. That’s a 7x improvement. Vectorization is king in pandas, no kidding.
Case 3: Smart Chunking + Resource Balancing
Problem: My chunks were uneven. Some processes had massive data loads; others were idle.
Fix: I wrote a dynamic chunk manager that split PAN-TAN groups more intelligently based on available memory and CPU cores. Now, multiple small chunks could be handled per process without starving or overloading the system.
# Here, we finalize the chunk size and decide how many tasks will run in parallel, aiming for as balanced resource usage as possible. .... .... .... grouped_task = [] temp_df1 = None temp_df2 = None total_rows_df1 = sum(groups_df1[k].shape[0] for k in common_keys) total_rows_df2 = sum(groups_df2[k].shape[0] for k in common_keys) total_rows = max(total_rows_df1, total_rows_df2) chunk_size = min(MIN_CHUCK_SIZE, total_rows // max_workers) for key in common_keys: if temp_df1 is None: temp_df1 = groups_df1.pop(key) temp_df2 = groups_df2.pop(key) else: temp_df1 = pd.concat([temp_df1, groups_df1.pop(key)], ignore_index=True) temp_df2 = pd.concat([temp_df2, groups_df2.pop(key)], ignore_index=True) if temp_df1.shape[0]>=chunk_size or temp_df2.shape[0]>=chunk_size: grouped_task.append((shared_list, key, temp_df1, temp_df2, ..., ...., ....)) temp_df1 = None temp_df2 = None if temp_df1 is not None: grouped_task.append((shared_list, key, temp_df1, temp_df2, ..., ..., ....)) temp_df1 = None temp_df2 = None .... .... ....
Result: Execution time fell to just 6 minutes. Yes, from 48 hours to 6 minutes. And no fancy tools -- just pandas, multiprocessing, and smart engineering.
Conclusion
- Avoid the use of .apply() unless you really have to. It’s intuitive, but slow for big data.
- Chunk your data smartly – think in groups, especially if your business logic allows for clean separation (like PAN-TAN in my case).
- Don’t over-parallelize – multiprocessing is powerful, but only when you balance memory, I/O, and CPU smartly.
- Merge related logic where possible to avoid redundant data loads
- Vectorized code isn’t just faster – it’s cleaner and easier to maintain.

